There are no projects in the garbage can.
Genomics
The Genomics unit focuses on the characterisation of the genome, the relationship between its variants, its expression and its regulation. Using different approaches, it studies the structure of our genetic material, its function and evolution. As well as the integration of the effect of other genetic and environmental elements on the expression of its genes.
In order to be able to carry out these studies, we have fine-tuned and developed workflows that allow us to detect SNPs, CNVs and Indels, changes in genome expression and in the regulation of this expression. Within the latter field, we have developed techniques that allow us to evaluate the content of regulatory RNAs: lncRNAs, miRNAs, snRNAs, piRNAs. Methods that allow us to evaluate modifications in DNA bases (methylation and acetylation) and the interaction of DNA with different proteins.
Thanks to the collaboration with the centre’s research groups and regular collaborators from the University of Granada and the Regional Ministry of Health, we have made progress in areas such as pharmacogenetics, liquid biopsy, characterisation of individualised cell types, determination of microvesicles, capture of regions in the genome and metagenomic studies. We have adapted these studies to a wide range of health areas: oncology, rare diseases, cardiovascular, inflammatory, mental, endocrine; as well as in many cases evaluating their application to daily clinical practice.
Members
Scientific Responsible
Technical Responsible
Research Support Technicians: Genomics Unit
Research Support Technicians: Bioinformatics Unit
Service Portfolio
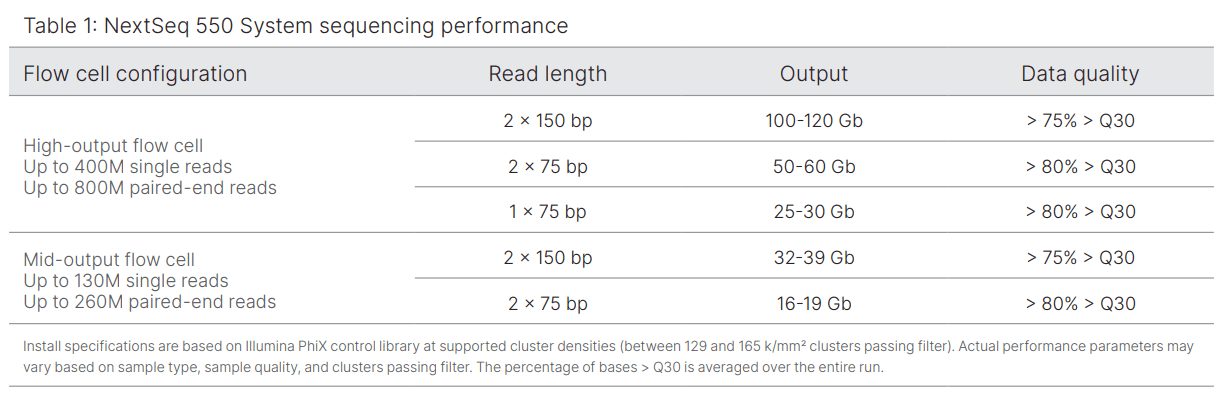
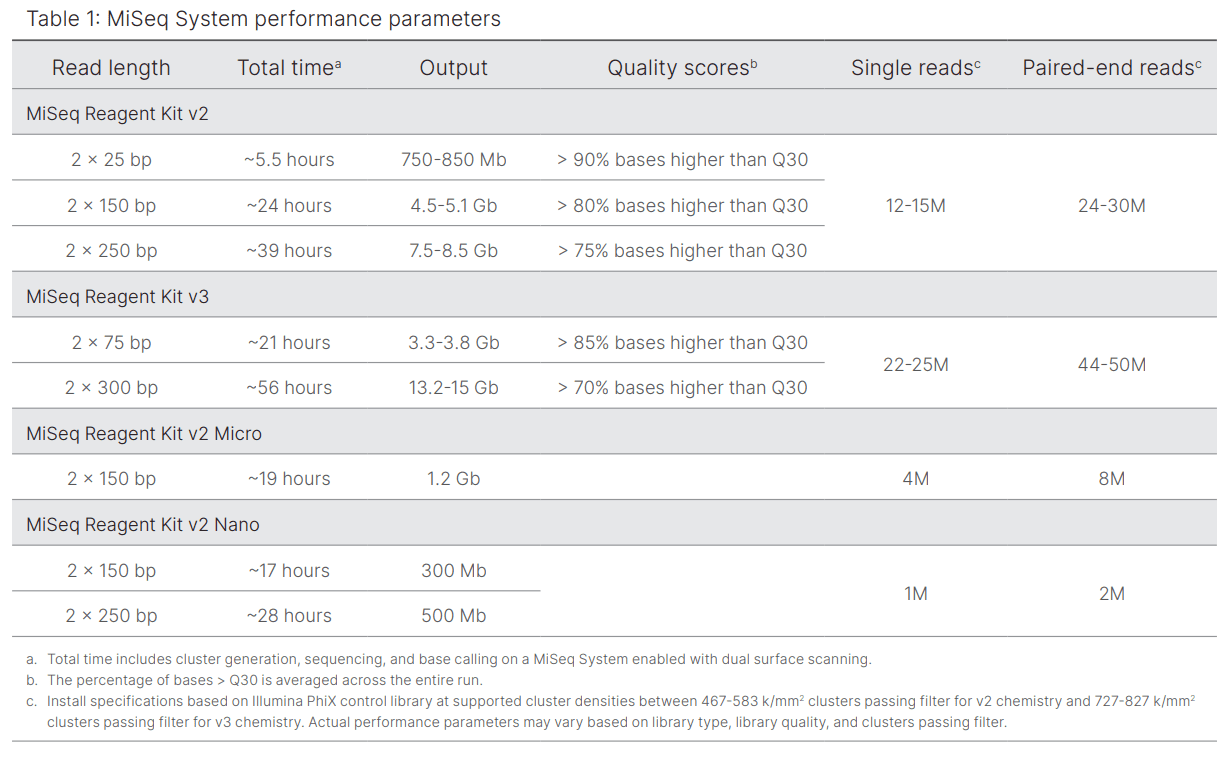
Equipment
Bioinformatics
Services:
The unit offers consultancy and bioinformatics analysis services covering both standard analyses of the most frequent experimental designs for the main massive sequencing platforms and arrays and ad-hoc analyses in research projects. These analyses include, among others:
- Design of experiments.
- Statistical analysis.
- Use of software and databases in bioinformatics.
- Mentoring of young researchers.
- Consultancy
Transcriptome and methylome analysis.
Standard analysis includes the following steps:
- Quality Control.
- Pre-processing and Normalisation.
- Statistical analysis (differential expression, clustering, classification, time series analysis).
- Functional analysis.
Genotyping Arrays (snps Arrays)
- Quality Control.
- Raw data processing.
- Normalisation.
- SNP and GWAS detection.
- CNV.
Genome and exome analysis
In targeted DNA and whole genome sequencing studies we provide analysis services including:
- Quality control on raw data.
- Alignment against reference genome.
- Variant identification (SNPS, InDels, etc.).
- Variant annotation and filtering.
Equipment
Compute node, equipped with 2 Intel Xeon Silver 4214R processors with 12 cores each at 2.4 GHz, with a total of 24 cores and 48 processing threads, and equipped with 256 GB of RAM and 10 TB of disk storage.
General Information
The main mission of GENyO’s Bioinformatics Unit is to assist and help researchers, both internal and external, in the process of handling, analysing and interpreting data obtained from next-generation sequencing experiments. To this end, we have expert staff and the necessary software and hardware resources to offer high quality services in the handling and processing of genomic data.
Over the last decade, the so-called omics technologies have brought about a profound revolution in biomedical research. Methods such as massive sequencing, DNA microarrays or proteomics and metabolomics techniques are allowing researchers to study cellular mechanisms from a global perspective and are proving to be extremely useful for characterising the molecular processes associated with different pathologies.
These new technologies are generating enormous amounts of data, and their storage, processing and interpretation require the use of advanced statistical and data mining techniques, as well as the use of high-performance computational resources for efficient data management. It is in this context that Bioinformatics has become an essential discipline in all genomics research centres.
GENyO’s Bioinformatics Unit focuses its activity on the development and application of new methods for the integrated analysis of omics data through the analysis and interpretation of data using statistical and computational techniques in internal and external projects.