Genomics Unit
General Information
The Genomics and Genotyping Unit of GENYO is dedicated mainly to high-throughput SNP genotyping, DNA sequencing, and gene expression quantification.
The services available in the Unit are exclusively aimed at research projects. Services of the Unit are available to all the researchers that request them, with preference to the research staff of GENYO and collaborators, and to the projects that follow the preferential research lines of the Center.
GENYO has the initiative of building partnerships with other Research Centers and Companies, and is interested in providing the best service to the field of research, therefore GENYO offers to the users reduced rates and processing times.
The Units of GENYO want to offer an excellent research setting focused on patients, with continuous improvements and the best quality of results. We provide the most efficient and adequate materials and methods. The scientific staff of the Unit, largely experienced on genomics, will provide support to users about the best technology to be applied in each project. Time of performance and rates will be adjusted to each project, without detriment to the quality of results.
The budget and working plan of the projects submitted to the Unit will be examined by the scientific staff and will be calculated based on prior works, according to the technical difficulties, issues and repetitions necessary from the beginning, avoiding biased results. It also will depend on the relationship of the applicant researcher with the Center and the work overload of the Unit.

Professor Marta Alarcón Riquelme is the Scientific Director of the Unit. She leads several research groups. She is largely experienced in molecular genetic research. Since the beginning of her scientific career, she has frequently used sequencing and genotyping platforms.
Thanks to the use of these technologies it has been possible to describe several susceptibility genes in the disease pathogenesis responsible for several complex diseases such as systemic lupus erythematosus, rheumatoid arthritis, psoriasis, etc.
Her studies are aimed not only at describing genes or polymorphisms associated to these diseases, but also the relationship between polymorphisms and gene expression mechanisms and their changes on coding proteins. Her group has also described the possible genetic interactions between genes associated to diseases.
Her experience with the new high-throughput technologies and the results obtained has promoted numerous national and international collaborations thanks to the groups involved and to the samples collected. All this has made possible that Professor Alarcón and her group are leaders in the study of genetic diseases associated with groups of population.
Technical Staff of the Unit
Contact
List of Services
 |
 |

Rt-PCR
An overview of these products can be found below:
Placas TaqMan® (96-384 Arrays). Más de 270 placas prediseñadas, algunas de ellas:
Pre-designed TaqMan® plates and products (384-well Micro fluidic Cards)
This product (TaqMan® Custom Array 384-well micro fluidic card) is able to perform up to 384 Taqman assays without robot, and analyze between 1 and 8 samples.
Some of the plates are:
Additional information of these assays and other products are available at Applied Biosystems by life technologiesTM: Real-time PCR
Sequencing by capillary electrophoresis
Automated DNA sequencing by Applied Biosystems uses the Sanger method. Once sequencing reaction is set up, the product is separated by capillary electrophoresis.
This technology enables to perform gene analysis with a high rate of mutation by the use of primers designed and optimized by the user as well as mitochondrial DNA study. Small (BigDye 1.1) 75 to 400pb fragments, and medium (BigDye 3.1) up to 1000pb fragments can be sequenced.
Complete gene amplicon sequence (RSA´s). VariantSEQr™ technology by M13 tailed primers.
Identification of microorganisms by 16S ribosomal sequencing.
Applications based on fragments analysis:
| Basic Service: Only sequencing. |
Full Service: Every effort to get results. |
|
| Verification, quality, and concentration | NO | YES |
| Concentration adjustment | NO | YES |
| Sequencing reaction and electrophoresis | YES | YES |
| Visualization, edition, and assessment of the quality of results | NO | YES |
| Report of results by email | YES | YES |
| Free repetition of the sequence | NO | YES |
| Delivery time | 3 working days | 5 working days |
Additional information of these assays and other products are available at Applied Biosystems by life technologiesTM: Real-time PCR
High-throughput genotyping
Illumina BeadArray technology: it includes a wide range of types of arrays, and a large list of SNPs for inclusion on the arrays, including SNPs designed by the researcher.
For this type of studies, we recommend to contact the Project Manager of the Service. Your Project Manager will help ensure all parts involved on the design of your project and best suit your requirements. Send us a list of SNPs you want to analyze and we will confirm you the availability to be performed with an Illumina assay technology.
Some of the products from Illumina can be found below:
|
Basic Service:
Only genotyping. |
Full Service:
Higher exploitation of results. |
|
|
Support on the experiment ‘s design
|
NO
|
YES
|
|
Verification, quality, and concentration
|
NO
|
YES
|
|
Concentration adjustment
|
NO
|
YES
|
|
Genomic amplification, enzymatic treatment, marking array
|
YES
|
YES
|
|
Report of results
|
YES
|
YES
|
|
Visualization, edition ,and assessment of the quality of results
|
NO
|
YES
|
|
Preliminary biostatistical analysis
|
NO
|
YES
|
|
Recording and backup of results
|
NO
|
YES
|
Next Generation Sequencing
SOLiD sequencing (This system will be available at very short notice).
All the services will be updated according to your requirements. Please contact us.
Our initial services include:
Complete exome resequencing and directed resequencing; whole exome resequencing, response of phenotype to drugs, medicines, or disease involved specific genes, detection of structural alterations and mutations.
Throughput Microarrays Scanners Systems
 |
 |
Next Generation Sequencing
Plataforma NextSeq 500, by Illumina. Plataforma de secuenciación de nueva generación para secuenciación masiva de ADN:
Permite la secuenciación de genomas completos, exomas y transcriptomas con muy alto rendimiento. La tecnología líder a nivel mundial, gracias a la química “Sequencing By Synthesis” (SBS):
5500xl SOLID System, from Applied Biosystems by Life Technologies. It is a genetic analysis platform for massive DNA sequencing.
This system is used for mega, advanced or massive sequencing (deep-sequencing) studies. It allows users to perform complete sequences above DNA 1 base G, using multiplexing system. It also allows to perform sequencing of complete small genomes identification of the complete genetic variability, measurement of transcripts in specific samples, and epigenetic studies; a more accurate technology improving reading, clarifying domains and variables, in GC-rich and AT-rich regions; full automated preparation of samples, with minimum quantity of errors. This system reduces costs importantly and compare the information obtained and the total cost (GB/€).
This equipment allows to perform the following studies:

 |
 |
Capillary Electrophoresis Sequencing
3130 HID Genetic analyzer, by Applied Biosystems by Life Technologies, for the analysis of DNA fragments, SNPs genotyping, and DNA sequencing.

Quantification systems of gene expression and real-time PCR genotyping
Two platforms TaqMan 7900HT, by Applied Biosystems, Life Technologies. Real-time PCR systems with capacity of 1-30 positions analysis in 384 samples. Higher accuracy equipment, with laser excitation source for scanning.
The 7500 Fast Real-Time PCR System, by Applied Biosystems, Life Technologies.
 |
 |
The QuantStudio™ 12K Flex System with OpenArray™ system. It is a highly flexible, real-time PCR platform that enables a broad range of applications with maximum throughput to researchers. This efficient and accurate system with OpenArray™ technology enables the scaling from 1 to 12,000 data points in a single run. It can produce up to 110.000 data points in a day. The components are designed to increase throughput and efficiency, reducing the time of manipulation, and costs.

The system includes 11 formats in order to provide flexible solutions in the plates configuration for the analysis of gene expression, miRNA or genotyping.
| Expression analysis | Samples/Plate | Genotyping Analysis | Samples/Plate |
| 18 (3X) assays | 48 | 16 assays | 144 |
| 56 assays | 48 | 32 assays | 96 |
| 112 assays | 24 | 64 assays | 48 |
| 168 assays | 16 | 128 assays | 24 |
| 224 assays | 12 | 192 assays | 16 |
| 256 assays | 12 |
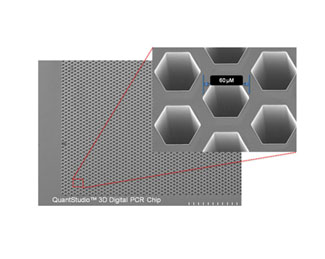
Digital PCR
The QuantStudio™ 3D Digital PCR System, is a simple and affordable platform that allows quantification and detection of specific sequences of nucleid acids by endpoint method. After amplification it enables to obtain up to 20,000 data points per sample, which facilitates the absolute quantification of gene expression, copy number variation (CNV) study, and low-frequency events.
 |
 |
Other equipment available

Links
The sequencing, genotyping and analysis of expression and regulation enable the identification of genes associated with physiological and pathological processes. These techniques provide information that makes possible the development of new early detection systems, the assessment of the genetic predisposition to have a disease, and the development of new and more efficient drugs to combat complicated diseases.
Sequencing
DNA sequencing is a technique used to study the genetic information of individuals. It is able to determine the order of nucleotides within DNA channels. Different methods have been developed to obtain the precise sequence of DNA nucleotides. The predominant methods used nowadays for DNA sequencing have been the enzymatic method and automated sequencing based on the Sanger method, which made possible the development of genomics within the last 3 decades.
A new generation of sequencing technologies, different from the Sanger method, has revolutionized the field of genomics, increasing enormously the velocity and the information provided.
Today, the new technologies have implemented the conventional techniques, such as sequencing by synthesis together with pyrosequencing, polymerase, or ligation based- sequencing.
In this field, the scientific board of GENYO had decided to provide the Genomics and Genotyping Unit with SOLID next-generation sequencing technology (Applied Biosystems by Life Technologies), based on the fragments generation and sequencing by ligation. This Unit also provides researchers with multicapillary electrophoresis systems.
 |
 |
Genotyping
Genotyping is the process of determining the genetic information of an organism- called genotype- and be able to differentiate it from the rest.
We understand genotyping as the analysis of one or several regions or positions. This technique was traditionally based on the study of the change of a specific position-nucleotide- which is called SNP, of a region or a repeat fragment (depending on their size: VNTR, STR, …). There are several techniques that allow to examine one position alone or millions positions.
There are different types of genotyping:
The Genotyping Unit covers all the possible demands of the researchers, providing different types of technologies and all the kits or microarrays pre-designed by the best trade houses. Some of these technologies are BeadArray™ (Illumina®) and Targeted Resequencing, SNPlex™, Taqman™,… (Applied Biosystems by Life Technologies™), among others.
Although genotyping can be addressed from different platforms, real Time-PCR, capillary or massive sequencing, DNA matrices, (arrays or biochips)… , the microarrays Illumina® make possible the use of BeadArray™ (Infinium and GoldenGate) technology for SNPs analysis, in the analysis of variability selected by the researcher. This system has several products available such as the Infinium iSelect HD Custom Genotyping BeadChip which allows the study of SNPs and structural variations. The array is offered on a 24-sample and 12-sample BeadChip (24-sample format with 3,000 to 68,000 analysis, or the 12-sample format with 68,000 to 200,000 positions).. Illumina offers a semi-custom product called the Human1M-Duo DNA Analysis BeadChip; it is a biochip that allows researchers to analyze two samples, with nearly 1.2 million markers per sample; of them, more than 60,800 SNPs are of the user’s choice. On the other hand, there are other biochips designed to Whole-Genome Genotyping studies or biochips specific for gene association studies, SNPs, regions of interest… of a particular disease, such as cáncer, using the GoldenGate technology.
Expression and regulation
Although cells of the organism all have the same genetic content, each one has a different phenotype. Cellular differentiation depends on the genome expression and regulation. We define expression and regulation as the molecular processes by which information encoded in an organism’s DNA is expressed. Each gene must be expressed at the right time and in the right place for a correct cellular function. To know the function of genes, we must know when, where, and how they are expressed. Changes in gene expression patterns provide information about the regulatory mechanisms, cell functions and metabolic pathways. To know the regulation of gene expression and its interactions provides information about diseases of different complexity such as cancer, neurodegenerative,or autoimmune diseases.
Gene expression analysis and large-scale methylation studies can be carried out using next-generation sequencers or high-density oligonucleotide microarray technology. However, today one of the most used techniques providing other complementary information is the real-time or quantitative PCR, which is able to detect the increase in the number of copies of DNA fragments in a cycle of PCR.
Our Unit has chosen Applied Biosystems by life technologies, based on the detection of fluorescence combined with PCR reaction. For the detection of PCR product, two types of chemistry are used: SYBR Green fluorochrome, and TaqMan specific fluorescent probes.
 |
 |
The Genomics Unit has been partially funded by ERDF funds (FEDER)



 Intranet
Intranet